Current methodologies, such as genome-wide associations (GWAS), miss so many SNPS that they cannot predict response for most GxEs.
Genteract’s new genetic discovery methodologies find a much greater fraction of the SNPs in each GxE and generates accurate GxE predictions.
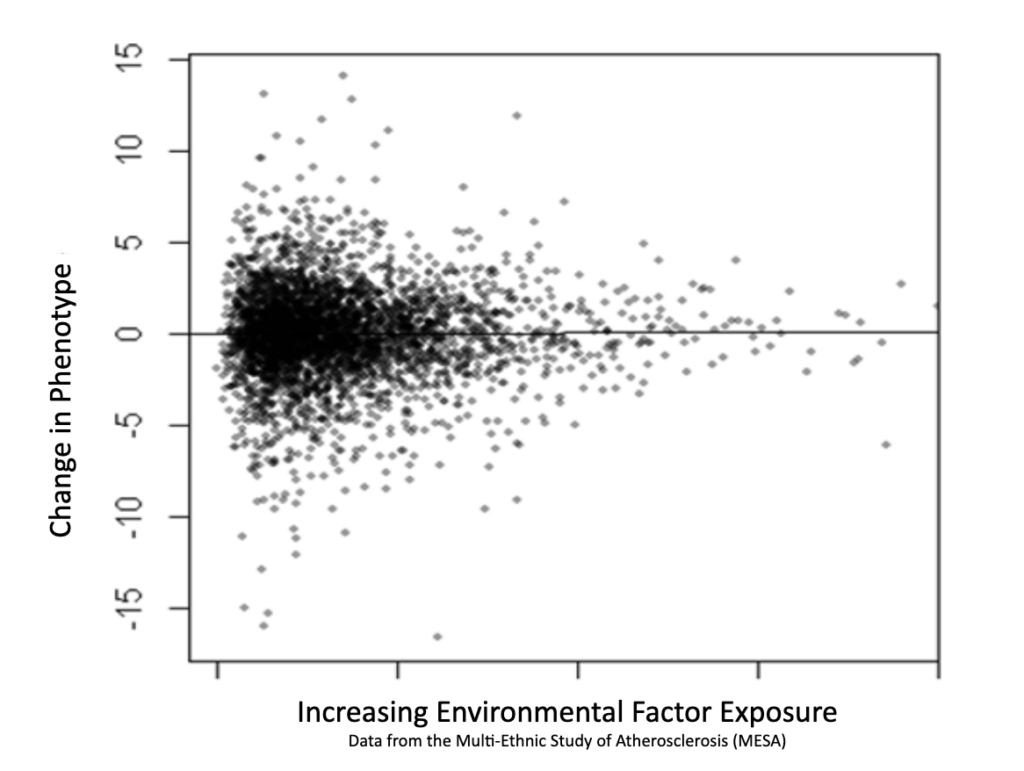
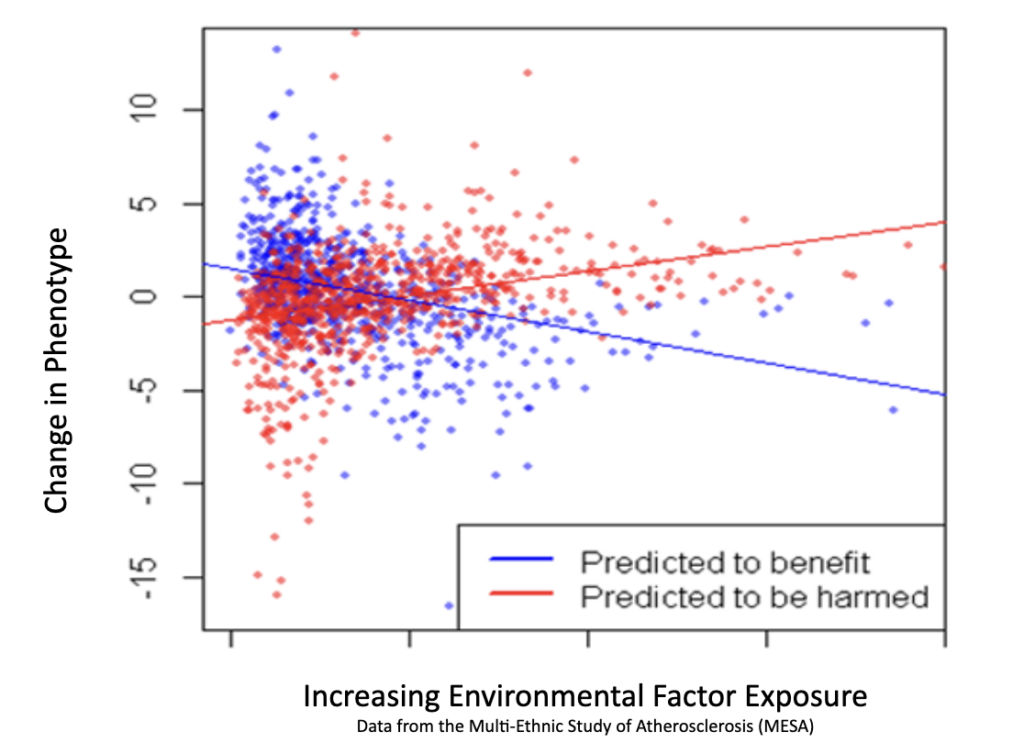
We use the output of our patented genetic discovery method, along with medical data to train an AI foundation model to make accurate biomedical predictions.
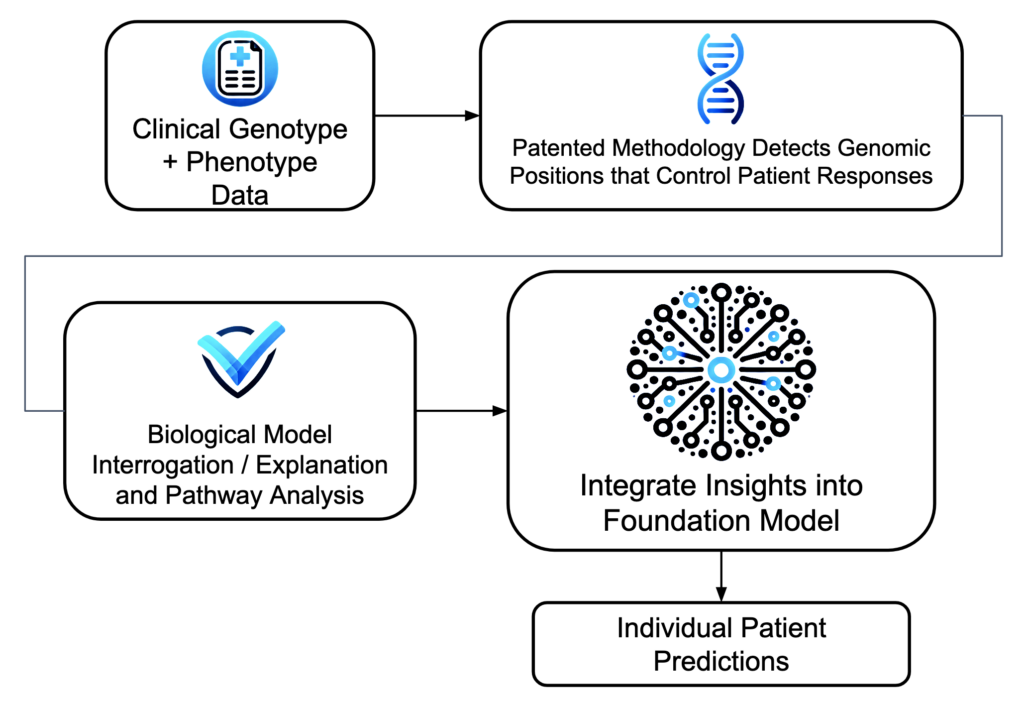
Insights learned from each clinical dataset are portable– they improve foundation model performance for every subsequent predictive task because each dataset increases the model’s understanding of human biology.
Deliver cost savings for payers and improved quality of care by predicting which drugs will work the best, with the fewest side effects, for each patient.
Identify genetically-defined patient subgroups who respond differently to a drug, including new indications for existing drugs.
Rescue drugs rejected by the FDA in phase III due to lack of broad efficacy, through a test that identifies patients who are strong responders.
Determine a patient's risk of diseases and find pharmaceutical and nutritional interventions to mitigate that risk.
The clinical trial or EMR data you already have may have massive hidden value that Genteract Analysis can unlock.
Contact us now to discuss a no-obligation analysis of your data to create highly accurate GxE predictions with cross-validation.
